🧬 1.5 생물학적 서열의 기본 처리
1.5 생물학적 서열의 기본 처리
🧬 이번에는 앞서서 만든 여러 함수들을 가지고 DNA 서열을 받으면 기본적으로 처리해야 할 것이 무엇이 있는지 정리해보자.
1.5.1. 기본적 처리 요약
🧬 다음 코드를 위해서는 앞선 포스팅에서 구현한 함수들이 sequences.py 라는 파일에 모두 들어가 있어야 한다.
🧬 1. validate_dna( ) : seq 서열의 DNA 유효성을 검사
🧬 2. transcription( ) : DNA 서열을 전사한 RNA 서열 생성
🧬 3. reverse_complement( ) : 역상보서열 생성
🧬 4. gc_content( ) : DNA 서열에서 GC 염기의 비율 확인
🧬 5. translate_seq( ) : DNA 서열을 아미노산 서열로 번역
🧬 6. all_orfs_ord( ) : DNA 서열을 단백질 서열로 변환
from sequences import *
seq = input("Insert DNA sequence: ")
if validate_dna(seq):
print("Valid sequence")
print("Transcription: ", transcription(seq))
print("Reverse complement: ", reverse_complement(seq))
print("GC content: ", gc_content(seq))
print("Direct translation: ", translate_seq(seq))
print("All proteins in ORFs(decreasing size): ", all_orfs_ord(seq))
else:
print("DNA sequence is not valid")
>>
Insert DNA sequence: ATGGGATCGTAGTCGTACTAGCTAGCTGATGGTACTCGATAGTCTACGTAGCTAGTGGTACTGGATGGTACTCAGTAACAT
Valid sequence
Transcription: AUGGGAUCGUAGUCGUACUAGCUAGCUGAUGGUACUCGAUAGUCUACGUAGCUAGUGGUACUGGAUGGUACUCAGUAACAU
Reverse complement: ATGTTACTGAGTACCATCCAGTACCACTAGCTACGTAGACTATCGAGTACCATCAGCTAGCTAGTACGACTACGATCCCAT
GC content: 0.4567901234567901
Direct translation: MGS_SY_LADGTR_ST_LVVLDGTQ_H
All proteins in ORFs(decreasing size): ['MLLSTIQYH', 'MVLDSLRS', 'MGS']
1.5.2. 파일을 읽고 쓰기
🧬 read_seq_from_file( ) : 주어진 파일을 읽기 모드로 불러와서 여러 줄에 있는 내용을 한 줄로 읽어들임 - \n 을 일반간격으로 replace 해서 읽음
🧬 DNA sequence read.txt 라는 파일에 미리 세 줄의 DNA서열을 입력해 두었다.

def read_seq_from_file(filename):
fh = open(filename, "r")
lines = fh.readlines()
seq=""
for l in lines:
seq += l.replace("\n", "")
fh.close()
return seq
print(read_seq_from_file('DNA sequence read.txt'))
>> ATGGGATCGTAGTCGTACTAGCTAGCTGATGGTACTCGATAGTCTACGTAGCTAGTGGTACTGGATGGTACTCAGTAACAT
DNA sequence read.txt 파일의 서열을 읽어온 것을 확인할 수 있다.
🧬 write_seq_to_file( ) : 주어진 파일을 쓰기 모드로 불러오거나 파일을 생성해서 텍스트 파일에 내용을 작성
def write_seq_to_file(seq, filename):
fh = open(filename, "w")
fh.write(seq)
fh.close()
return None
write_seq_to_file("ATGGGATCGTAGTCGTACTAGCTAGCTGATGGTACTCGATAGTCTACGTAGCTAGTGGTACTGGATGGTACTCAGTAACAT", 'DNA sequence write.txt')
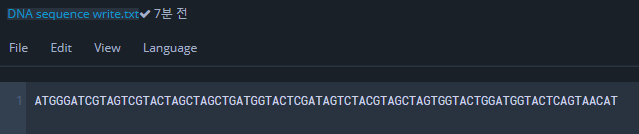
DNA sequence write.txt 파일에 서열이 입력되는 것을 확인하자.
1.5.3. DNA의 최종 기본적 처리
🧬 .txt 파일의 DNA 서열을 read_seq_from_file( ) 함수로 읽어옴
🧬 읽어온 DNA 서열에서 최종적으로 얻고자 하는 것은 결국 발현되는 단백질이기 때문에 이 단백질 서열을 파일에 작성해서 최종 처리함
🧬 all_orfs_ord( ) 함수를 호출하여 모든 리딩프레임에 대해서 개시코돈과 종결코돈을 고려한 단백질만 가져온다.
🧬 all_orfs_ord( ) 함수에서 얻은 단백질 서열을 write_seq_to_file( ) 함수가 orf-i.txt 이름으로 생성한 파일에 작성해줌
from sequences import *
fname = input("Insert input filename: ")
seq = read_seq_from_file(fname)
if validate_dna(seq):
print("Valid sequence")
print("Transcription: ", transcription(seq))
print("Reverse complement: ", reverse_complement(seq))
print("GC content: ", gc_content(seq))
print("Direct translation: ", translate_seq(seq))
orfs = all_orfs_ord(seq)
i = 1
for orf in orfs:
write_seq_to_file(orf, "orf-"+ str(i) + ".txt")
i += 1
else:
print("DNA sequence is not valid")
>>
Insert input filename: DNA sequence read.txt
Valid sequence
Transcription: AUGGGAUCGUAGUCGUACUAGCUAGCUGAUGGUACUCGAUAGUCUACGUAGCUAGUGGUACUGGAUGGUACUCAGUAACAU
Reverse complement: ATGTTACTGAGTACCATCCAGTACCACTAGCTACGTAGACTATCGAGTACCATCAGCTAGCTAGTACGACTACGATCCCAT
GC content: 0.4567901234567901
Direct translation: MGS_SY_LADGTR_ST_LVVLDGTQ_H

위 사진에서 확인할 수 있듯이 orf-i.txt 파일이 생성되었다.

각 파일에 단백질 서열이 크기순서대로 입력된 것을 확인할 수 있다.
1.5.4. 요약
🧬 위의 함수들을 통해서 우리가 직접 서열을 입력할 수도 있지만 특정 파일에 있는 서열을 읽어오는 방법도 배웠다. 특히 유전 서열은 그 길이나 크기가 굉장히 크기 때문에 일일이 입력하기보다는 읽어오는 것이 더 편하다고 생각한다.
🧬 이렇게 DNA 서열을 받아오면 우리는 우선 서열의 유효성을 검사하고 이를 전사한 RNA 서열을 만들어본다. 그리고 서열의 GC 염기 비율을 알아보고, 이 서열이 주형가닥인지 비주형가닥인지 확인이 힘든 경우에는 역상보서열을 만들어서 DNA 서열을 아미노산 서열로 번역한다. 마지막으로 이 아미노산 서열을 단백질 서열로 반환하면 DNA 서열을 가지고 할 수 있는 전처리는 어느정도 끝났다고 할 수 있다.
Leave a comment